基本概念
交叉: 变量间的双向影响(A -> B 和 B -> A)
滞后: 时间上的先后关系(过去 -> 未来)
用于解决因果方向问题, 解决”先有鸡还是先有蛋”的争议.
- 单向因果关系: 若仅仅一方路径显著(如 BMI -> 肝功能), 则支持单向因果假设(如:肥胖导致肝损伤). 至少需要两个时间点的观测数据
模型结构:
自回归路径: 同一变量在不同时间点的稳定性 (如时间1的BMI预测时间2的BMI)
交叉滞后路径: 不同变量间跨时间影响 (如时间1的BMI预测时间2的肝功能, 反之亦然)
同步协方差: 同一时间点不同变量之间的关联 (如时间1的BMI与肝功能的相关性)
正系数: 同一时间点的变量值同向变化
负系数: 同一时间点的反向变化
Demo
#数据制备---------
library(foreign)
library(haven)
library(dplyr)
data <- read_dta("data/charls/H_CHARLS_D_Data.dta")
harmonized<-dplyr::select(data,ID,inw1,inw3,
ragender,r1agey,r3agey,r1smokev,r3smokev,
r1drinkev,r3drinkev,r1diabe,r3diabe,
r1mbmi,r3mbmi,r1livere,r3livere)
harmonized <- subset(harmonized, inw1 == 1&inw3 == 1)
summary(harmonized)
ID inw1 inw3 ragender r1agey
Length:14576 Min. :1 Min. :1 Min. :1.000 Min. : 10.00
Class :character 1st Qu.:1 1st Qu.:1 1st Qu.:1.000 1st Qu.: 50.00
Mode :character Median :1 Median :1 Median :2.000 Median : 57.00
Mean :1 Mean :1 Mean :1.528 Mean : 57.97
3rd Qu.:1 3rd Qu.:1 3rd Qu.:2.000 3rd Qu.: 64.00
Max. :1 Max. :1 Max. :2.000 Max. :101.00
NA's :38
r3agey r1smokev r3smokev r1drinkev
Min. : 14.00 Min. :0.0000 Min. :0.0000 Min. :0.0000
1st Qu.: 54.00 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:0.0000
Median : 61.00 Median :0.0000 Median :0.0000 Median :0.0000
Mean : 61.95 Mean :0.3902 Mean :0.4453 Mean :0.3886
3rd Qu.: 68.00 3rd Qu.:1.0000 3rd Qu.:1.0000 3rd Qu.:1.0000
Max. :105.00 Max. :1.0000 Max. :1.0000 Max. :1.0000
NA's :38 NA's :82 NA's :54 NA's :98
r3drinkev r1diabe r3diabe r1mbmi
Min. :0.0000 Min. :0.00000 Min. :0.00000 Min. : 0.00
1st Qu.:0.0000 1st Qu.:0.00000 1st Qu.:0.00000 1st Qu.: 20.85
Median :0.0000 Median :0.00000 Median :0.00000 Median : 23.12
Mean :0.4546 Mean :0.05812 Mean :0.09994 Mean : 24.08
3rd Qu.:1.0000 3rd Qu.:0.00000 3rd Qu.:0.00000 3rd Qu.: 25.77
Max. :1.0000 Max. :1.00000 Max. :1.00000 Max. :2449.95
NA's :76 NA's :209 NA's :448 NA's :2854
r3mbmi r1livere r3livere
Min. : 12.73 Min. :0.00000 Min. :0.0000
1st Qu.: 21.08 1st Qu.:0.00000 1st Qu.:0.0000
Median : 23.51 Median :0.00000 Median :0.0000
Mean : 24.59 Mean :0.03447 Mean :0.0674
3rd Qu.: 26.12 3rd Qu.:0.00000 3rd Qu.:0.0000
Max. :2366.09 Max. :1.00000 Max. :1.0000
NA's :2870 NA's :186 NA's :347
harmonized <- na.omit(harmonized)
names(harmonized)
[1] "ID" "inw1" "inw3" "ragender" "r1agey" "r3agey"
[7] "r1smokev" "r3smokev" "r1drinkev" "r3drinkev" "r1diabe" "r3diabe"
[13] "r1mbmi" "r3mbmi" "r1livere" "r3livere"
# 模型定义与拟合----------
# 交叉滞后模型定义与拟合
#交叉
library(tidySEM)
library(lavaan)
This is lavaan 0.6-19
lavaan is FREE software! Please report any bugs.
#AI指令:拟合BMI与肝病的交叉滞后模型,校正协变量:。。。。
cross_lag_model <- '
# 自回归路径
r3mbmi ~ r1mbmi
r3livere ~ r1livere
# 交叉滞后路径
r3mbmi ~ r1livere
r3livere ~ r1mbmi
# 校正协变量 (人口学、生活方式、疾病史)
r3mbmi ~ ragender + r3agey + r3smokev + r3drinkev + r3diabe
r3livere ~ ragender + r3agey + r3smokev + r3drinkev + r3diabe
# Wave1同时点相关
r1mbmi ~~ r1livere
r1mbmi ~~ ragender + r1agey + r1smokev + r1drinkev + r1diabe
r1livere ~~ ragender + r1agey + r1smokev + r1drinkev + r1diabe
# Wave3同时点残差相关
r3mbmi ~~ r3livere
'
fit <- sem(cross_lag_model, data=harmonized, estimator="MLR")
# 输出模型拟合结果(这里的示例模型拟合很差,仅供教学)
#CFI >0.90
#TLI >0.90
#RMSEA <0.08
#SRMR <0.05
summary(fit, standardized = TRUE, fit.measures = TRUE)
lavaan 0.6-19 ended normally after 123 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 35
Number of observations 9433
Model Test User Model:
Standard Scaled
Test Statistic 107327.706 61996.699
Degrees of freedom 46 46
P-value (Chi-square) 0.000 0.000
Scaling correction factor 1.731
Yuan-Bentler correction (Mplus variant)
Model Test Baseline Model:
Test statistic 113904.804 72458.861
Degrees of freedom 72 72
P-value 0.000 0.000
Scaling correction factor 1.572
User Model versus Baseline Model:
Comparative Fit Index (CFI) 0.058 0.144
Tucker-Lewis Index (TLI) -0.475 -0.340
Robust Comparative Fit Index (CFI) 0.058
Robust Tucker-Lewis Index (TLI) -0.475
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -139244.863 -139244.863
Scaling correction factor 127.906
for the MLR correction
Loglikelihood unrestricted model (H1) NA NA
Scaling correction factor 56.251
for the MLR correction
Akaike (AIC) 278559.726 278559.726
Bayesian (BIC) 278810.045 278810.045
Sample-size adjusted Bayesian (SABIC) 278698.821 278698.821
Root Mean Square Error of Approximation:
RMSEA 0.497 0.378
90 Percent confidence interval - lower 0.495 0.376
90 Percent confidence interval - upper 0.500 0.380
P-value H_0: RMSEA <= 0.050 0.000 0.000
P-value H_0: RMSEA >= 0.080 1.000 1.000
Robust RMSEA 0.497
90 Percent confidence interval - lower 0.494
90 Percent confidence interval - upper 0.500
P-value H_0: Robust RMSEA <= 0.050 0.000
P-value H_0: Robust RMSEA >= 0.080 1.000
Standardized Root Mean Square Residual:
SRMR 0.234 0.234
Parameter Estimates:
Standard errors Sandwich
Information bread Observed
Observed information based on Hessian
Regressions:
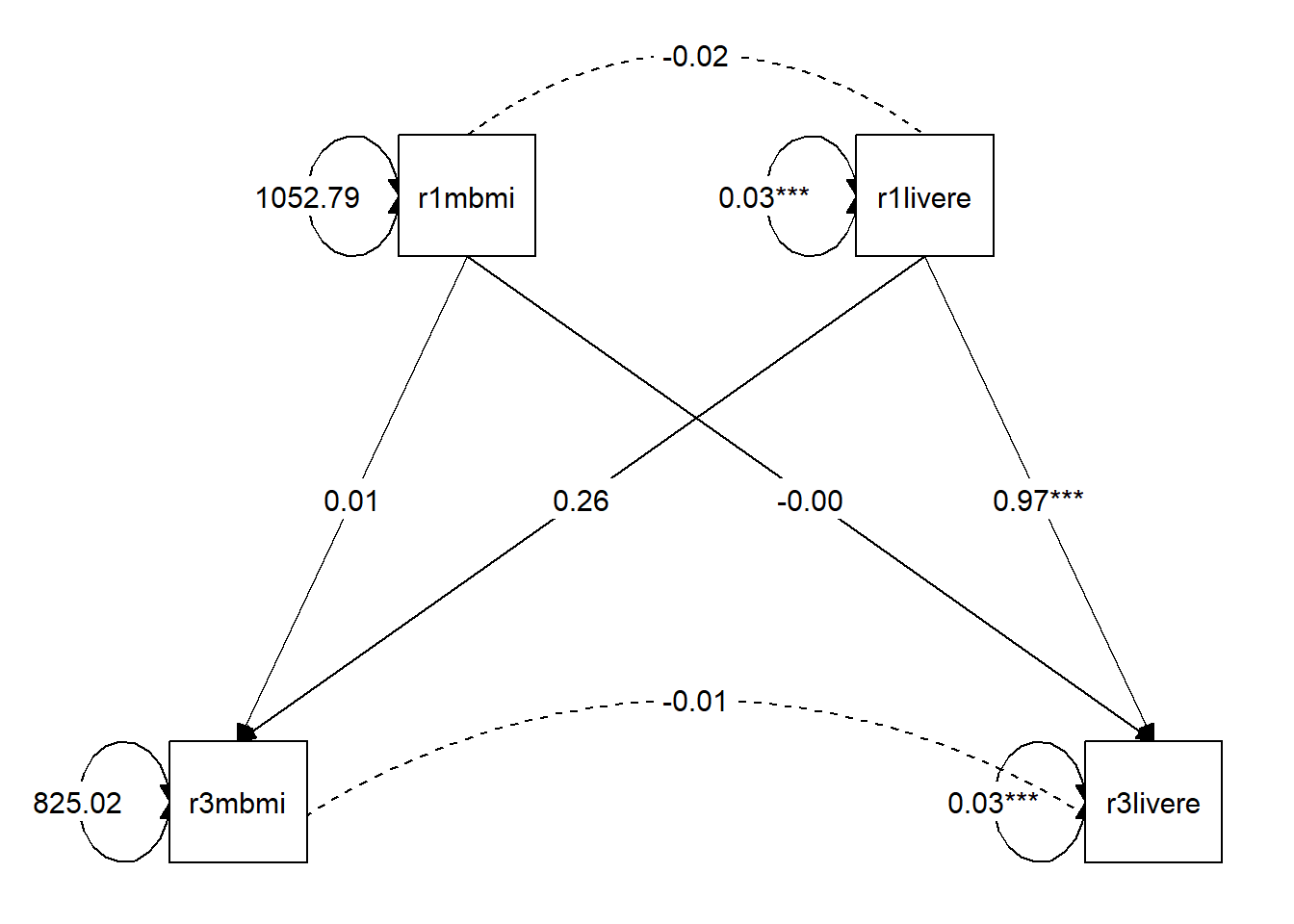
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
r3mbmi ~
r1mbmi 0.010 0.007 1.553 0.120 0.010 0.012
r3livere ~
r1livere 0.966 0.002 460.069 0.000 0.966 0.705
r3mbmi ~
r1livere 0.256 0.765 0.334 0.738 0.256 0.002
r3livere ~
r1mbmi -0.000 0.000 -1.289 0.198 -0.000 -0.002
r3mbmi ~
ragender 0.823 0.422 1.948 0.051 0.823 0.014
r3agey -0.064 0.018 -3.563 0.000 -0.064 -0.021
r3smokev -1.065 0.322 -3.305 0.001 -1.065 -0.018
r3drinkev -0.292 0.567 -0.515 0.607 -0.292 -0.005
r3diabe 0.742 0.459 1.617 0.106 0.742 0.008
r3livere ~
ragender -0.002 0.006 -0.310 0.756 -0.002 -0.004
r3agey -0.000 0.000 -1.926 0.054 -0.000 -0.012
r3smokev -0.000 0.006 -0.015 0.988 -0.000 -0.000
r3drinkev 0.004 0.004 0.913 0.361 0.004 0.008
r3diabe 0.032 0.008 4.089 0.000 0.032 0.039
Covariances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
r1mbmi ~~
r1livere -0.016 0.013 -1.255 0.209 -0.016 -0.003
ragender 0.257 0.121 2.127 0.033 0.257 0.016
r1agey -3.698 5.221 -0.708 0.479 -3.698 -0.012
r1smokev -0.134 0.108 -1.246 0.213 -0.134 -0.009
r1drinkev -0.034 0.077 -0.445 0.656 -0.034 -0.002
r1diabe 0.077 0.024 3.167 0.002 0.077 0.010
r1livere ~~
ragender -0.001 0.001 -0.854 0.393 -0.001 -0.014
r1agey -0.017 0.017 -0.982 0.326 -0.017 -0.010
r1smokev 0.000 0.001 0.247 0.805 0.000 0.004
r1drinkev 0.001 0.001 1.158 0.247 0.001 0.014
r1diabe 0.001 0.001 1.610 0.107 0.001 0.019
.r3mbmi ~~
.r3livere -0.006 0.011 -0.597 0.550 -0.006 -0.001
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.r3mbmi 825.017 588.383 1.402 0.161 825.017 0.999
.r3livere 0.030 0.002 18.039 0.000 0.030 0.501
r1mbmi 1052.789 689.262 1.527 0.127 1052.789 1.000
r1livere 0.032 0.002 18.570 0.000 0.032 1.000
ragender 0.248 0.000 555.841 0.000 0.248 1.000
r1agey 86.834 1.273 68.222 0.000 86.834 1.000
r1smokev 0.236 0.001 201.799 0.000 0.236 1.000
r1drinkev 0.235 0.001 193.783 0.000 0.235 1.000
r1diabe 0.054 0.002 25.508 0.000 0.054 1.000
#路径图-------------
# 定义布局
lay <- get_layout(
"", "r1mbmi", "", "r1livere", "",
"", "", "", "", "",
"r3mbmi", "", "", "", "r3livere",
rows = 3)
# 绘制路径图(不包含协变量)
graph_sem(fit, layout=lay, angle=170)
Some edges involve nodes not in layout. These were dropped.